Please see the slides for a more detailed explanation.
DEAP Short Tutorial
DEAP (Distributed Evolutionary Algorithms for Python) is a Python module which provides you with tools to easily implement evolutionary computation algorithms.
In this short tutorial you’ll learn how to set up everything to start working with it right away.
First off, install and import as you would do with any Python module.
from deap import base, creator, tools # deap utilities
import random
import numpy as np # numerical computation
import matplotlib.pyplot as plt # plotting
Now we start by setting information about the problem:
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", list, fitness=creator.FitnessMin)
I’m creating two classes here:
- An
Individualwhich inherits thelisttype, with an attribute namedfitnesswhich is another object:- A
FitnessMinclass which inheritsbase.Fitness, with an atribute namedweightsinitialized at(-1.0,)
- A
Minimisation and maximisation are pretty similar in optimisation: you usually flip the symbol $<$ for $>$ which is setting the weight to -1 or 1.
Now let’s add some more info about the algorithm:
IND_SIZE = 10 # chromosome length
toolbox = base.Toolbox()
toolbox.register("attribute", random.random)
toolbox.register("individual", tools.initRepeat, creator.Individual,
toolbox.attribute, n=IND_SIZE)
toolbox.register("population", tools.initRepeat, list,toolbox.individual)
First we instantiate a Toolbox and register methods in it.
For example, we are registering the attribute method which is just calling random.random. It is generating an attribute (a gene), which in this case is a random float between 0 and 1.
The tools.initRepeat method is tricky. Taking a look at its documentation shows that it repeats a method $n$ times, and fills a container with it. On line 5, we are repeating toolbox.attribute IND_SIZE times, and filling the creator.Individual container. This process is registered now as toolbox.individual.
We now add the objective function and operators:
def evaluate(ind):
""" Returns the fitness of an individual.
This is your objective function."""
return sum(ind), # must return a tuple
# Operators
toolbox.register("cross", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutGaussian, mu=0, sigma=1, indpb=0.1)
toolbox.register("select", tools.selTournament, tournsize=3)
toolbox.register("evaluate", evaluate)
We now registered some of the operators dealing with the individuals.
As you can see, tools.cxTwoPoint is a two-point crossover. It receives two individuals and modify them inplace. A tuple is returned containing a view of two individuals.
The tools.mutGaussian, as you can guess, is the distribution of the mutation. This needs real-valued individuals of course.
Notice how on line 4 we return a tuple in the evaluate function, as fitness values must be iterables in order for multi-objective optimisation to be included in the schematics of the library.
Single objective optimisation is a special case of multi-objective: using 1-tuple.
Putting things together
This is the whole algorithm implemented using the tools we defined. It is a generational Genetic Algorithm—each generation will replace the previous one.
def generationalGA():
pop = toolbox.population(n=50) # Registered as a list
CXPB = 0.5 # Crossover probability
MUTPB = 0.2 # Mutation probability
NGEN = 40 # Number of generations
# Evaluate all population first
fitnesses = map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
# Generate offspring
for __ in range(NGEN):
offspring = toolbox.select(pop, len(pop))
offspring = list(map(toolbox.clone, offspring)) # Generate a deep copy
# Apply crossover
for c1, c2 in zip(offspring[::2], offspring[1::2]):
if random.random() < CXPB:
toolbox.cross(c1, c2)
# Reset their fitness values
del c1.fitness.values
del c2.fitness.values
# Mutate those selected
for mutant in offspring:
if random.random() < MUTPB:
toolbox.mutate(mutant)
# Reset fitness values
del mutant.fitness.values
# Evaluate non-evaluated individuals in offspring
invalid_inds = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = map(toolbox.evaluate, invalid_inds)
for ind, fit in zip(invalid_inds, fitnesses):
ind.fitness.values = fit
# Replace entire population by the offspring
pop[:] = offspring
return pop
Since we are getting a list as the return value, we can access each individual or its features accordingly:
results = generationalGA()
fitnesses = [i.fitness.getValues()[0] for i in results]
bestpos = fitnesses.index(min(fitnesses))
print("Individual: {0}\n\nFitness: {1}".format(
results[bestpos], results[bestpos].fitness.getValues()[0]))
Individual: [0.007255964641715962, -3.7128774659861112, -2.3940455890783134, -8.190571991822432, -0.8942735610322432, -8.585607468392073, -2.753605093491699, -2.015675322782945, -4.444005821030648, -5.351258705602769]
Fitness: -38.33466505457752
1 + 1 EA using NumPy arrays
As you probably noticed, we used plain old lists as the individual and population containers. We can use numpy arrays if you’re used to that, too. Let’s give it a go with the OneMax problem.
First problem information:
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("IndividualNP", np.ndarray, fitness=creator.FitnessMax)
The OneMax problem looks to maximise the number of ones in a chromosome, so this time we’re choosing from random integers between 0 and 1. The rest is pretty similar.
IND_LENGTH = 20
toolbox = base.Toolbox()
toolbox.register("attribute", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.IndividualNP,
toolbox.attribute, n=IND_LENGTH)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
- Objective function: In case of the OneMax problem, we want to maximise the number of 1s so the sum of an individual represents its fitness.
- Mutation: we can flip the bits, so we use
UniformIntbetween 0 and 1. Each bit has $1/n$ probability of changing.
def evaluate_ind(a):
return a.sum(), # return a tuple
toolbox.register("mutate", tools.mutUniformInt, low=0, up=1,
indpb=1.0 / IND_LENGTH)
toolbox.register("evaluate", evaluate_ind)
We’re gonna introduce a couple of useful tools now:
- Hall of Fame: Remember the best individual and its fitness over the entire timespan.
- Logbook: Record statistics of your algorithm to review later.
# Instantiate HoF and Logbook
hof = tools.HallOfFame(1, similar=np.array_equal) # Notice the np.array_equal
log = tools.Logbook()
def oneplusone():
# We don't really need a container, but let's
# stick to the original metaphor of evolution
pop = toolbox.population(n=1)
NGEN = 25 # Number of generations
for gen in range(NGEN):
parent = pop[0] # This is just an alias
# Evaluate everyone (i.e. one individual)
ind_fitness = list(map(toolbox.evaluate, pop))
# ...and set its fitness
parent.fitness.values = ind_fitness[0]
# Clone
child = toolbox.clone(parent)
# Always mutate
toolbox.mutate(child)
# ...and reset its fitness after mutation
del child.fitness.values
# Reevaluate the child
child.fitness.values = toolbox.evaluate(child)
# Compare & keep best
if child.fitness >= parent.fitness:
pop[:] = [child]
hof.update(pop)
# Record statistics
log.record(agen=gen, ind=pop[0], fit=pop[0].fitness.getValues()[0])
return pop
results = oneplusone()
print("Individual: {0}\n\nFitness: {1}".format(
results[0], results[0].fitness.getValues()[0]))
Individual: [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
Fitness: 12.0
We can select specific statistics from the logbook, or pretty-print them all at once:
print(log)
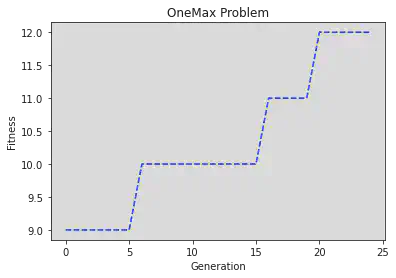
agen fit ind
0 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
1 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
2 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
3 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
4 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
5 9.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 0 0 0 1]
6 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
7 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
8 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
9 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
10 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
11 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
12 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
13 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
14 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
15 10.0 [1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1]
16 11.0 [1 1 0 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
17 11.0 [1 1 0 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
18 11.0 [1 1 0 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
19 11.0 [1 1 0 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
20 12.0 [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
21 12.0 [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
22 12.0 [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
23 12.0 [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
24 12.0 [1 1 1 0 0 1 1 0 1 1 0 1 1 0 0 1 1 0 0 1]
The statistics are already lists of values:
type(log.select("agen"))
list
So we’re able to easily plot them with pyplot:
gen = log.select("agen")
fit = log.select("fit")
fig, ax = plt.subplots()
line = ax.plot(gen, fit, "b--", label="Maximum Fitness")
ax.set_xlabel("Generation")
ax.set_ylabel("Fitness")
plt.title("OneMax Problem")
plt.show()